Introduction
PISA is a text search engine able to run on large-scale collections of documents. It allows researchers to experiment with state-of-the-art techniques, allowing an ideal environment for rapid development.
Some features of PISA are listed below:
- Written in C++ for performance
- Indexing, parsing & sharding capabilities
- Many index compression methods supported
- Many query processing algorithms supported
- Implementation of document reordering
- Free and open-source with permissive license
Note
PISA is still in its unstable release, and no stability or backwards-compatibility is guaranteed with each new version. New features are constantly added, and contributions are welcome!
Requirements
Compilers
To compile PISA, you will need a compiler supporting the C++20 standard. Our continuous integration pipeline compiles PISA and runs tests in the following configurations:
- Linux:
- GCC, versions: 10, 11, 12, 13, 14
- Clang 16, 17, 18, 19
There are currently no MacOS builds running in CI because we have no one actively maintaining them. That said, Mac builds should in theory work on the x86 architecture. The new ARM-based architecture has not been tested.
Supporting Windows is planned but is currently not being actively worked on, mostly due to a combination of man-hour shortage, prioritization, and no core contributors working on Windows at the moment. If you want to help us set up a Github workflow for Windows and work out some issues with compilation, let us know on our Slack channel.
System Dependencies
Most build dependencies are managed automatically with CMake and git submodules. However, several dependencies still need to be manually provided:
CMake >= 3.0autoconf,automake,libtool, andm4(for buildinggumbo-parser)- OpenMP (optional)
You can opt in to use some system dependencies instead of those in git submodules:
- Google Benchmark
(
PISA_SYSTEM_GOOGLE_BENCHMARK): this is a dependency used only for compiling and running microbenchmarks. - oneTBB (
PISA_SYSTEM_ONETBB): both build-time and runtime dependency. - Boost (
PISA_SYSTEM_BOOST): both build-time and runtime dependency. - CLI11 (
PISA_SYSTEM_CLI11): build-time only dependency used in command line tools.
For example, to use all the system installation of Boost in your build:
cmake -DPISA_SYSTEM_BOOST=ON <source-dir>
Installation
The following steps explain how to build PISA. First, you need the code checked out from Github. (Alternatively, you can download the tarball and unpack it on your local machine.)
$ git clone https://github.com/pisa-engine/pisa.git
$ cd pisa
Then create a build environment.
$ mkdir build
$ cd build
Finally, configure with CMake and compile:
$ cmake ..
$ make
Build Types
There are two build types available:
Release(default)DebugRelWithDebInfoMinSizeRel
Use Debug only for development, testing, and debugging. It is much slower at runtime.
Learn more from CMake documentation.
Build Systems
CMake supports configuring for different build systems. On Linux and Mac, the default is Makefiles, thus, the following two commands are equivalent:
$ cmake -G ..
$ cmake -G "Unix Makefiles" ..
Alternatively to Makefiles, you can configure the project to use Ninja instead:
$ cmake -G Ninja ..
$ ninja # instead of make
Other build systems should work in theory but are not tested.
Testing
You can run the unit and integration tests with:
$ ctest
The directory test/test_data contains a small document collection used in the
unit tests. The binary format of the collection is described in a following
section.
An example set of queries can also be found in test/test_data/queries.
Indexing Pipeline
This section is an overview of how to take a collection to a state in which it can be queried. This process is intentionally broken down into several steps, with a bunch of independent tools performing different tasks. This is because we want the flexibility of experimenting with each individual step without recomputing the entire pipeline.
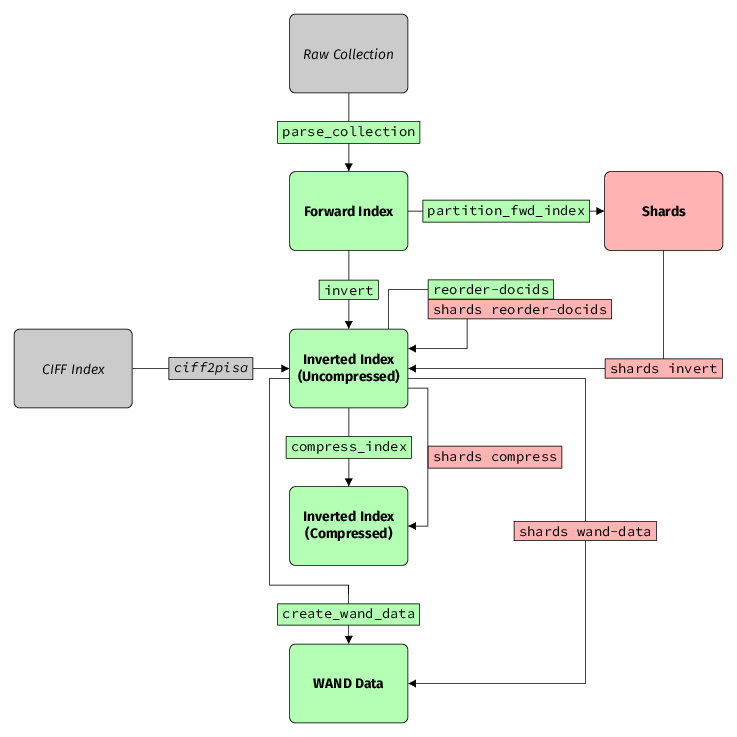
External Resources
Raw Collection
The raw collection is a dataset containing the documents to index. A
collection is encoded in one of the supported
formats that stores a list of document
contents along with some metadata, such as URL and title. The
parse_collection tool takes a collection as an input and parses it to
a forward index (see Forward Index). See
Parsing for more details.
CIFF Index
This is an inverted index in the Common Index File Format.
It can be converted to an uncompressed PISA index (more information below)
with the ciff2pisa tool.
Forward Index
A forward index is the output of the parse_collection tool.
It represents each document as a list of tokens (terms) in the order of their appearance.
To learn more about parsing and the forward index format, see Parsing.
Inverted Index
An inverted index is the most fundamental structure in PISA. For each term in the collection, it contains a list of documents the term appears in. PISA distinguishes two types of inverted index.
Uncompressed / Binary Collection
The uncompressed index stores document IDs and frequencies as 4-byte integers.
It is an intermediate format between forward index and compressed inverted index.
It is obtained by running invert on a forward index.
To learn more about inverting a forward index, see Inverting.
Optionally, documents can be reordered with reorder-docids to obtain another
instance of uncompressed inverted index with different assignment of IDs to documents.
More on reordering can be found in Document Reordering.
Compressed
An uncompressed index is large and therefore before running queries, it must be compressed with one of many available encoding methods. It is this compressed index format that is directly used when issuing queries. See Compress Index to learn more.
WAND Data
This is a special metadata file containing additional statistics used during query processing. See Build additional data.
Shards
PISA supports partitioning a forward index into subsets called shards.
Structures of all shards can be transformed in bulk using shards command line tool.
To learn more, read Sharding.
Parsing
A forward index is a data structure that stores the term identifiers associated to every document. Conversely, an inverted index stores for each unique term the document identifiers where it appears (usually, associated to a numeric value used for ranking purposes such as the raw frequency of the term within the document).
The objective of the parsing process is to represent a given collection
as a forward index. To parse a collection, use the parse_collection
command, for example:
$ mkdir -p path/to/forward
$ zcat ClueWeb09B/*/*.warc.gz | \ # pass unzipped stream in WARC format
parse_collection \
-j 8 \ # use up to 8 threads at a time
-b 10000 \ # one thread builds up to 10k documents in memory
-f warc \ # use WARC
-F lowercase porter2 \ # lowercase and stem every term (using the Porter2 algorithm)
--html \ # strip HTML markup before extracting tokens
-o path/to/forward/cw09b
In case you get the error -bash: /bin/zcat: Argument list too long,
you can pass the unzipped stream using:
$ find ClueWeb09B -name '*.warc.gz' -exec zcat -q {} \;
The parsing process will write the following files:
cw09b: forward index in binary format.cw09b.terms: a new-line-delimited list of sorted terms, where term having ID N is on line N, with N starting from 0.cw09b.termlex: a binary representation (lexicon) of the.termsfile that is used to look up term identifiers at query time.cw09b.documents: a new-line-delimited list of document titles (e.g., TREC-IDs), where document having ID N is on line N, with N starting from 0.cw09b.doclex: a binary representation of the.documentsfile that is used to look up document identifiers at query time.cw09b.urls: a new-line-delimited list of URLs, where URL having ID N is on line N, with N starting from 0. Also, keep in mind that each ID corresponds with an ID of thecw09b.documentsfile.
Generating mapping files
Once the forward index has been generated, a binary document map and
lexicon file will be automatically built. However, they can also be
built using the lexicon utility by providing the new-line delimited
file as input. The lexicon utility also allows efficient look-ups and
dumping of these binary mapping files.
For example, assume we have the following plaintext, new-line delimited
file, example.terms:
aaa
bbb
def
zzz
We can generate a lexicon as follows:
./bin/lexicon build example.terms example.lex
You can dump the binary lexicon back to a plaintext representation:
./bin/lexicon print example.lex
It should output:
aaa
bbb
def
zzz
You can retrieve the term with a given identifier:
./bin/lexicon lookup example.lex 2
Which outputs:
def
Finally, you can retrieve the id of a given term:
./bin/lexicon rlookup example.lex def
It outputs:
2
NOTE: This requires the initial file to be lexicographically sorted,
as rlookup uses binary search for reverse lookups.
Supported stemmers
Both are English stemmers. Unfortunately, PISA does not have support for any other languages. Contributions are welcome.
Supported formats
The following raw collection formats are supported:
plaintext: every line contains the document's title first, then any number of whitespaces, followed by the content delimited by a new line character.jsonl: every line is a JSON document with three fields:title,content, and (optionally)url(NOTE: "title" in PISA is a unique string identifying a document)trectext: TREC newswire collections.trecweb: TREC web collections.warc: Web ARChive format as defined in the format specification.wapo: TREC Washington Post Corpus.
In case you want to parse a set of files where each one is a document (for example, the collection
wiki-large), use the files2trec.py script
to format it to TREC (take into account that each relative file path is used as the document ID).
Once the file is generated, parse it with the parse_collection command specifying the trectext
value for the --format option.
IR Datasets
We provide a convenient integration with
ir-datasets through a Python script that can
be piped to the parse_collection tool.
NOTE: the script depends on a Python 3 environment that has the
ir-datasets package already installed. See the project's documentation
for the installation instructions.
The script is called ir-datasets and is copied to the bin directory
when compiling the project. It takes one argument, which is the name of
the collection you want to parse. Please refer to
ir-datasets for the list of supported
datasets and for the instructions on how to connect the datasets that
are not downloaded directly, but rather have to be linked manually.
The documents are printed to the standard output in the JSONL format.
You can pipe the output of the ir-datasets script to parse_dataset
program, and pass --format jsonl to the latter.
$ ir-datasets wikir/en1k | \
parse_collection -f jsonl -F lowercase porter2 -o path/to/forward/index
Note that the first run of the ir-datasets script on a given
collection may take a while because it may have to download the data to
the local drive first.
Inverting
Once the parsing phase is complete, use the invert command to turn a
forward index into an inverted index. For example, assuming the
existence of a forward index in the path path/to/forward/cw09b:
$ mkdir -p path/to/inverted
$ ./invert -i path/to/forward/cw09b \
-o path/to/inverted/cw09b
Inverting an index requires the knowledge of the number of terms in
the lexicon ahead of time. In the above example, the invert command
assumes that a cw09b.termlex' file exists from the output of parse_collection` which is used to lookup the term count.
Note that the number of terms can be provided using --term-count in
case the lexicon is not available or on a different path.
Inverted index format
A binary sequence is a sequence of integers prefixed by its length,
where both the sequence integers and the length are written as 32-bit
little-endian unsigned integers. An inverted index consists of 3
files, <basename>.docs, <basename>.freqs, <basename>.sizes:
-
<basename>.docsstarts with a singleton binary sequence where its only integer is the number of documents in the collection. It is then followed by one binary sequence for each posting list, in order of term-ids. Each posting list contains the sequence of document-ids containing the term. -
<basename>.freqsis composed of a one binary sequence per posting list, where each sequence contains the occurrence counts of the postings, aligned with the previous file (note however that this file does not have an additional singleton list at its beginning). -
<basename>.sizesis composed of a single binary sequence whose length is the same as the number of documents in the collection, and the i-th element of the sequence is the size (number of terms) of the i-th document.
Reading the inverted index using Python
Here is an example of a Python script reading the uncompressed inverted index format:
import os
import numpy as np
class InvertedIndex:
def __init__(self, index_name):
index_dir = os.path.join(index_name)
self.docs = np.memmap(index_name + ".docs", dtype=np.uint32,
mode='r')
self.freqs = np.memmap(index_name + ".freqs", dtype=np.uint32,
mode='r')
def __iter__(self):
i = 2
while i < len(self.docs):
size = self.docs[i]
yield (self.docs[i+1:size+i+1], self.freqs[i-1:size+i-1])
i += size+1
def __next__(self):
return self
for i, (docs, freqs) in enumerate(InvertedIndex("cw09b")):
print(i, docs, freqs)
Compressing
To create an index use the command compress_inverted_index. The
available index types are listed in index_types.hpp.
For example, to create an index using the optimal partitioning algorithm, using the test collection, execute the command:
$ ./bin/compress_inverted_index -t opt \
-c ../test/test_data/test_collection \
-o test_collection.index.opt \
--check
where test/test_data/test_collection is the basename of the
collection, that is the name without the .{docs,freqs,sizes}
extensions, and test_collection.index.opt is the filename of the
output index. --check will trigger a verification step to check the
correctness of the index.
Compression Algorithms
Binary Interpolative Coding
Binary Interpolative Coding (BIC) directly encodes a monotonically increasing sequence. At each step of this recursive algorithm, the middle element m is encoded by a number m − l − p, where l is the lowest value and p is the position of m in the currently encoded sequence. Then we recursively encode the values to the left and right of m. BIC encodings are very space-efficient, particularly on clustered data; however, decoding is relatively slow.
To compress an index using BIC use the index type block_interpolative.
Alistair Moffat, Lang Stuiver: Binary Interpolative Coding for Effective Index Compression. Inf. Retr. 3(1): 25-47 (2000)
Elias-Fano
Given a monotonically increasing integer sequence S of size n, such that \(S_{n-1} < u\), we can encode it in binary using \(\lceil\log u\rceil\) bits. Elias-Fano coding splits each number into two parts, a low part consisting of \(l = \lceil\log \frac{u}{n}\rceil\) right-most bits, and a high part consisting of the remaining \(\lceil\log u\rceil - l\) left-most bits. The low parts are explicitly written in binary for all numbers, in a single stream of bits. The high parts are compressed by writing, in negative-unary form, the gaps between the high parts of consecutive numbers.
To compress an index using Elias-Fano use the index type ef.
Sebastiano Vigna. 2013. Quasi-succinct indices. In Proceedings of the sixth ACM international conference on Web search and data mining (WSDM ‘13). ACM, New York, NY, USA, 83-92.
MaskedVByte
Jeff Plaisance, Nathan Kurz, Daniel Lemire, Vectorized VByte Decoding, International Symposium on Web Algorithms 2015, 2015.
OptPFD
Hao Yan, Shuai Ding, and Torsten Suel. 2009. Inverted index compression and query processing with optimized document ordering. In Proceedings of the 18th international conference on World wide web (WWW '09). ACM, New York, NY, USA, 401-410. DOI: https://doi.org/10.1145/1526709.1526764
Partitioned Elias Fano
Giuseppe Ottaviano and Rossano Venturini. 2014. Partitioned Elias-Fano indexes. In Proceedings of the 37th international ACM SIGIR conference on Research & development in information retrieval (SIGIR '14). ACM, New York, NY, USA, 273-282. DOI: https://doi.org/10.1145/2600428.2609615
QMX
Quantities, Multipliers, and eXtractor (QMX) packs as many integers as possible into 128-bit words (Quantities) and stores the selectors (eXtractors) separately in a different stream. The selectors are compressed (Multipliers) with RLE (Run-Length Encoding).
To compress an index using QMX use the index type block_qmx.
Andrew Trotman. 2014. Compression, SIMD, and Postings Lists. In Proceedings of the 2014 Australasian Document Computing Symposium (ADCS '14), J. Shane Culpepper, Laurence Park, and Guido Zuccon (Eds.). ACM, New York, NY, USA, Pages 50, 8 pages. DOI: https://doi.org/10.1145/2682862.2682870
SIMD-BP128
Daniel Lemire, Leonid Boytsov: Decoding billions of integers per second through vectorization. Softw., Pract. Exper. 45(1): 1-29 (2015)
Simple8b
Vo Ngoc Anh, Alistair Moffat: Index compression using 64-bit words. Softw., Pract. Exper. 40(2): 131-147 (2010)
Simple16
Jiangong Zhang, Xiaohui Long, and Torsten Suel. 2008. Performance of compressed inverted list caching in search engines. In Proceedings of the 17th international conference on World Wide Web (WWW '08). ACM, New York, NY, USA, 387-396. DOI: https://doi.org/10.1145/1367497.1367550
StreamVByte
Daniel Lemire, Nathan Kurz, Christoph Rupp: Stream VByte: Faster byte-oriented integer compression. Inf. Process. Lett. 130: 1-6 (2018). DOI: https://doi.org/10.1016/j.ipl.2017.09.011
Varint-G8IU
Alexander A. Stepanov, Anil R. Gangolli, Daniel E. Rose, Ryan J. Ernst, and Paramjit S. Oberoi. 2011. SIMD-based decoding of posting lists. In Proceedings of the 20th ACM international conference on Information and knowledge management (CIKM '11), Bettina Berendt, Arjen de Vries, Wenfei Fan, Craig Macdonald, Iadh Ounis, and Ian Ruthven (Eds.). ACM, New York, NY, USA, 317-326. DOI: https://doi.org/10.1145/2063576.2063627
VarintGB
Jeffrey Dean. 2009. Challenges in building large-scale information retrieval systems: invited talk. In Proceedings of the Second ACM International Conference on Web Search and Data Mining (WSDM '09), Ricardo Baeza-Yates, Paolo Boldi, Berthier Ribeiro-Neto, and B. Barla Cambazoglu (Eds.). ACM, New York, NY, USA, 1-1. DOI: http://dx.doi.org/10.1145/1498759.1498761
"WAND" Data
WARNING
"WAND data" is a legacy term that may be somewhat misleading. The name originates from the WAND algorithm, but in fact all scored queries must use WAND data. This name will likely change in the future to more accurately reflect its nature, but we still use it now because it is prevalent throughout the code base.
This is a file that contains data necessary for scoring documents, such as:
- document lengths,
- term occurrence counts,
- term posting counts,
- number of documents,
- term max scores,
- (optional) term block-max scores.
Use create_wand_data command to build.
Quantization
If you quantize your inverted index, then you have to quantize WAND data as well, using the same number of bits (otherwise, any algorithms that depend on max-scores will not function correctly).
Compression
You can build your WAND data with score compression. This will store scores quantized, thus you need to provide the number of quantization bits. Note that even if you compress your WAND data, you can still use it with a non-quantized index: the floating point scores will be calculated (though they will not be identical to the original scores, as this compression is lossy). If you do use a quantized index, it must use the same number of bits as WAND data.
Querying
The command queries treats each line of the standard input (or a file if -q
is present) as a separate query. A query line contains a whitespace-delimited
list of tokens. These tokens are either interpreted as terms (if --terms is
defined, which will be used to resolve term IDs) or as term IDs (if --terms is
not defined). Optionally, a query can contain query ID delimited by a colon:
Q1:one two three
^^ ^^^^^^^^^^^^^
query ID terms
For example:
$ ./bin/queries \
-e opt \ # index encoding
-a and \ # retrieval algorithm
-i test_collection.index.opt \ # index path
-w test_collection.wand \ # metadata file
-q ../test/test_data/queries # query input file
This performs conjunctive queries (and). In place of and other operators can
be used (see Query algorithms). To run multiple algorithms,
provide -a multiple times (for example, -a and -a or -a wand).
The tool outputs a JSON with query execution statistics including mean, median
(q50), and percentiles (q90, q95, q99) for different per-query
aggregation (none, min, mean, median, max).
If the WAND file is compressed, append --compressed-wand flag.
Supported algorithms
The following algorithms are available via the -a option:
andoror_freqwandblock_max_wandblock_max_maxscoreranked_andblock_max_ranked_andranked_ormaxscoreranked_or_taatranked_or_taat_lazy
Additional options
--runs <N>: Number of runs per query (default: 3)-o, --output <FILE>: Output file for per-run query timing data--safe: Rerun if not enough results with pruning (requires--thresholds)--quantized: Quantized scores
Build additional data
To perform BM25 queries it is necessary to build an additional file containing the parameters needed to compute the score, such as the document lengths. The file can be built with the following command:
$ ./bin/create_wand_data \
-c ../test/test_data/test_collection \
-o test_collection.wand
If you want to compress the file append --compress at the end of the command.
When using variable-sized blocks (for VBMW) via the --variable-block
parameter, you can also specify lambda with the -l <float> or
--lambda <float> flags. The value of lambda impacts the mean size of the
variable blocks that are output. See the VBMW paper (listed below) for more
details. If using fixed-sized blocks, which is the default, you can supply the
desired block size using the -b <UINT> or --block-size <UINT> arguments.
Retrieval Algorithms
This is the list of the supported query processing algorithms.
Unranked
PISA implements two unranked algorithms, meaning they return a full list of documents matching the query in the order of their appearance in the posting lists.
Intersection
The intersection algorithm (and) returns only the documents that match
all query terms.
Union
The union algorithm (or) returns all the documents that match any
query term.
Top-k Retrieval
Top-k retrieval returns the top-k highest scored documents with respect To the given query.
Document-at-a-time (DaaT)
Document-at-a-time algorithms traverse one document at a time. They rely on posting lists being sorted by document IDs, and scan them in step to retrieve all frequencies for a document right away.
Conjunctive processing
Conjunctive processing (ranked_and) returns the top k documents that
contain all of the query terms. This is an exhaustive algorithm,
meaning all documents must be scored.
Disjunctive processing
Conjunctive processing (ranked_or) returns the top k documents that
contain any of the query terms. This is an exhaustive algorithm,
meaning all documents must be scored.
MaxScore
MaxScore (maxscore) uses precomputed maximum partial scores for each
term to avoid calculating all scores. It is especially suitable for
longer queries (high term count), short posting lists, or high values of
k (number of returned top documents).
Howard Turtle and James Flood. 1995. Query evaluation: strategies and optimizations. Inf. Process. Manage. 31, 6 (November 1995), 831-850. DOI=http://dx.doi.org/10.1016/0306-4573(95)00020-H
WAND
Similar to MaxScore, WAND (wand) uses precomputed maximum partial
scores for each term to avoid calculating all scores. Its performance is
sensitive to the term count, so it may not be the best choice for long
queries. It may also take a performance hit when k is very high, in
which case MaxScore may prove more efficient.
Andrei Z. Broder, David Carmel, Michael Herscovici, Aya Soffer, and Jason Zien. 2003. Efficient query evaluation using a two-level retrieval process. In Proceedings of the twelfth international conference on Information and knowledge management (CIKM '03). ACM, New York, NY, USA, 426-434. DOI: https://doi.org/10.1145/956863.956944
BlockMax WAND
BlockMax WAND (block_max_wand) builds on top of WAND. It uses
additional precomputed scores for ranges of documents in posting lists,
which allows for skipping entire blocks of documents if their max score
is low enough.
Shuai Ding and Torsten Suel. 2011. Faster top-k document retrieval using block-max indexes. In Proceedings of the 34th international ACM SIGIR conference on Research and development in Information Retrieval (SIGIR '11). ACM, New York, NY, USA, 993-1002. DOI=http://dx.doi.org/10.1145/2009916.2010048
Variable BlockMax WAND
Variable BlockMax WAND is the same algorithm as block_max_wand at
query time. The difference is in precomputing the block-max scores.
Instead having even block sizes, each block can have a different size,
to optimize the effectiveness of skips.
Antonio Mallia, Giuseppe Ottaviano, Elia Porciani, Nicola Tonellotto, and Rossano Venturini. 2017. Faster BlockMax WAND with Variable-sized Blocks. In Proceedings of the 40th International ACM SIGIR Conference on Research and Development in Information Retrieval (SIGIR '17). ACM, New York, NY, USA, 625-634. DOI: https://doi.org/10.1145/3077136.3080780
BlockMax MaxScore
BlockMax MaxScore (block_max_maxscore) is a MaxScore implementation
with additional block-max scores, similar to BlockMax WAND.
BlockMax AND
BlockMax AND (block_max_ranked_and) is a conjunctive algorithm using
block-max scores.
Term-at-a-time (TaaT)
Term-at-a-time algorithms traverse one posting list at a time. Thus, they cannot rely on all frequencies for a given document being known at the time of their processing. This requires an accumulator structure to keep partial scores.
Disjunctive TaaT processing
Disjunctive TaaT (ranked_or_taat) is a simple algorithm that
accumulates document scores while traversing postings one list at a
time. ranked_or_taat_lazy is a variant that uses an accumulator array
that initializes lazily.
Document Reordering
PISA supports reassigning document IDs that were initially assigned in order of parsing. The point of doing it is usually to decrease the index size or speed up query processing. This part is done on an uncompressed inverted index. Depending on the method, you might also need access to some parts of the forward index. We support the following ways of reordering:
- random,
- by a feature (such as URL or document TREC ID),
- with a custom-defined mapping, and
- recursive graph bisection.
All of the above are supported by a single command reorder-docids.
Below, we explain each method and show some examples of running the command.
Reordering document lexicon
All methods can optionally take a path to a document lexicon and make a copy of it that reflects the produced reordering.
reorder-docids \
--documents /path/to/original/doclex \
--reordered-documents /path/to/reordered/doclex \
...
Typically, you will want to do that if you plan to evaluate queries, which will need access to a correct document lexicon.
NOTE: Because these options are common to all reordering methods, we ignore them below for brevity.
Random
Random document reordering, as the name suggests, randomly shuffles all document IDs. Additionally, it can take a random seed. Two executions of the command with the same seed will produce the same final ordering.
reorder-docids --random \
--collection /path/to/inv \
--output /path/to/inv.random \
--seed 123456789 # optional
By feature (e.g., URL or TRECID)
An index can be reordered according to any single document feature, such as URL or TRECID,
as long as it is stored in a text file line by line, where line n is the feature of
document n in the original order.
In particular, our collection parsing command produces two such feature files:
*.documents, which is typically a list of TRECIDs,*.urls, which is a list of document URLs.
To use either, you simply need to run:
reorder-docids \
--collection /path/to/inv \
--output /path/to/inv.random \
--by-feature /path/to/feature/file
From custom mapping
You can also produce a mapping yourself and feed it to the command. Such mapping is a text file with two columns separated by a whitespace:
<original ID> <new ID>
Having that, reordering is as simple as running:
reorder-docids \
--collection /path/to/inv \
--output /path/to/inv.random \
--from-mapping /path/to/custom/mapping
Recursive Graph Bisection
We provide an implementation of the Recursive Graph Bisection (aka BP) algorithm, which is currently the state-of-the-art for minimizing the compressed space used by an inverted index (or graph) through document reordering. The algorithm tries to minimize an objective function directly related to the number of bits needed to store a graph or an index using a delta-encoding scheme.
Learn more from the original paper:
L. Dhulipala, I. Kabiljo, B. Karrer, G. Ottaviano, S. Pupyrev, and A. Shalita. Compressing graphs and indexes with recursive graph bisection. In Proc. SIGKDD, pages 1535–1544, 2016.
In PISA, you simply need to pass --recursive-graph-bisection option (or its alias --bp)
to the reorder-docids command.
reorder-docids --bp \
--collection /path/to/inv \
--output /path/to/inv.random
Note that --bp allows for some additional options.
For example, the algorithm constructs a forward index in memory, which is in a special format
separate from the PISA forward index that you obtain from the parse_collection tool.
You can instruct reorder-docids to store that intermediate structure (--store-fwdidx),
as well as provide a previously constructed one (--fwdidx), which can be useful if you
want to reuse it for several runs with different algorithm parameters.
To see all available parameters, run reorder-docids --help.
Sharding
We support partitioning a collection into a number of smaller subsets called shards.
Right now, only a forward index can be partitioned by running partition_fwd_index command.
For convenience, we provide shards command that supports certain bulk operations on all shards.
Partitioning collection
We support two methods of partitioning: random, and by a defined mapping. For example, one can partition collection randomly:
$ partition_fwd_index \
-j 8 \ # use up to 8 threads at a time
-i full_index_prefix \
-o shard_prefix \
-r 123 # partition randomly into 123 shards
Alternatively, a set of files can be provided.
Let's assume we have a folder shard-titles with a set of text files.
Each file contains new-line-delimited document titles (e.g., TREC-IDs) for one partition.
Then, one would call:
$ partition_fwd_index \
-j 8 \ # use up to 8 threads at a time
-i full_index_prefix \
-o shard_prefix \
-s shard-titles/*
Note that the names of the files passed with -s will be ignored.
Instead, each shard will be assigned a numerical ID from 0 to N - 1 in order
in which they are passed in the command line.
Then, each resulting forward index will have appended .ID to its name prefix:
shard_prefix.000, shard_prefix.001, and so on.
Working with shards
The shards tool allows to perform some index operations in bulk on all shards at once.
At the moment, the following subcommands are supported:
- invert,
- compress,
- wand-data, and
- reorder-docids.
All input and output paths passed to the subcommands will be expanded for each individual shards
by extending it with .<shard-id> (e.g., .000) or, if substring {} is present, then
the shard number will be substituted there. For example:
shards reorder-docids --by-url \
-c inv \
-o inv.url \
--documents fwd.{}.doclex \
--reordered-documents fwd.url.{}.doclex
is equivalent to running the following command for every shard XYZ:
reorder-docids --by-url \
-c inv.XYZ \
-o inv.url.XYZ \
--documents fwd.XYZ.doclex \
--reordered-documents fwd.url.XYZ.doclex
Threshold Estimation
Currently it is possible to perform threshold estimation tasks using the
kth_threshold tool. The tool computes the k-highest impact score for
each term of a query. Clearly, the top-k threshold of a query can be
lower-bounded by the maximum of the k-th highest impact scores of the
query terms.
In addition to the k-th highest score for each individual term, it is possible to use the k-th highest score for certain pairs and triples of terms.
To perform threshold estimation use the kth_threshold command.
Regression test for Robust04
This tutorial explains how to run regression tests on the Robust04 collection.
Requirements
Due to the collection's license, we cannot distribute it, and thus you will need to obtain it separately to run this test. See https://trec.nist.gov/data/cd45/index.html for more information.
Docker Image
In the repository, we provide a Docker image definition under
test/docker/benchmark to make reproducing results easier. First, to
build the image, run the following command from the repository's root
directory:
docker build -t pisa-bench -f- .. < test/docker/benchmark/Dockerfile
You can use any name instead of pisa-bench but if you use a different
one, make sure to substitute it in any subsequent command.
Building the image may take a while because the tools will be compiled.
Once the image is built, you can run it with:
podman run \
-v path/to/disk45:/opt/disk45:ro \
-v your/workdir:/opt/workdir \
--rm pisa-bench
Replace path/to/disk45 with your local path to the Robust04 disk 4 and
5 directory, and your/workdir with the path to a local working
directory, where artifacts will be written.
By default, the script will build all indices and run evaluation. If you want to inspect the image or run some custom commands, you can execute the image interactively:
podman run \
-v $HOME/data/disk45:/opt/disk45:ro \
-v $HOME/workdir:/opt/workdir \
--rm -it pisa-bench /bin/bash
compress_inverted_index
Usage
Compresses an inverted index
Usage: ../../../build/bin/compress_inverted_index [OPTIONS]
Options:
-h,--help Print this help message and exit
-c,--collection TEXT REQUIRED
Uncompressed index basename
-o,--output TEXT REQUIRED Output inverted index
--check Check the correctness of the index
-e,--encoding TEXT REQUIRED Index encoding
-w,--wand TEXT Needs: --scorer
WAND data filename
-s,--scorer TEXT Needs: --wand --quantize
Scorer function
--bm25-k1 FLOAT Needs: --scorer
BM25 k1 parameter.
--bm25-b FLOAT Needs: --scorer
BM25 b parameter.
--pl2-c FLOAT Needs: --scorer
PL2 c parameter.
--qld-mu FLOAT Needs: --scorer
QLD mu parameter.
--quantize UINT Needs: --scorer
Quantizes the scores using this many bits
-L,--log-level TEXT:{critical,debug,err,info,off,trace,warn} [info]
Log level
--config Configuration .ini file
Description
Compresses an inverted index from the uncompressed format using one of the integer encodings.
Input
The input to this command is an uncompressed version of the inverted
index described here.
The --collection option takes the basename of the uncompressed
index.
Encoding
The postings are compressed using one of the available integer
encodings, defined by --encoding. The available encoding values are:
block_interpolative: Binary Interpolative Codingef: Elias-Fanoblock_maskedvbyte: MaskedVByteblock_optpfor: OptPForDeltapef: Partitioned Elias-Fanoblock_qmx: QMXblock_simdbp: SIMD-BP128block_simple8b: Simple8bblock_simple16: Simple16block_streamvbyte: StreamVByteblock_varintg8iu: Varint-G8IUblock_varintgb: Varint-GB
Precomputed Quantized Scores
At the time of compressing the index, you can replace frequencies with
quantized precomputed scores. To do so, you must define --quantize
flag, plus some additional options:
--scorer: scoring function that should be used in to calculate the scores (bm25,dph,pl2,qld)--wand: metadata filename path
compute_intersection
Usage
Computes intersections of posting lists.
Usage: ../../../build/bin/compute_intersection [OPTIONS]
Options:
-h,--help Print this help message and exit
-e,--encoding TEXT REQUIRED Index encoding
-i,--index TEXT REQUIRED Inverted index filename
-w,--wand TEXT REQUIRED WAND data filename
--compressed-wand Needs: --wand
Compressed WAND data file
--tokenizer TEXT:{english,whitespace} [english]
Tokenizer
-H,--html Strip HTML
-F,--token-filters TEXT:{krovetz,lowercase,porter2} ...
Token filters
--stopwords TEXT Path to file containing a list of stop words to filter out
-q,--queries TEXT Path to file with queries
--terms TEXT Term lexicon
--weighted Weights scores by query frequency
--min-query-len INT Minimum query length
--max-query-len INT Maximum query length
-s,--scorer TEXT REQUIRED Scorer function
--bm25-k1 FLOAT Needs: --scorer
BM25 k1 parameter.
--bm25-b FLOAT Needs: --scorer
BM25 b parameter.
--pl2-c FLOAT Needs: --scorer
PL2 c parameter.
--qld-mu FLOAT Needs: --scorer
QLD mu parameter.
-L,--log-level TEXT:{critical,debug,err,info,off,trace,warn} [info]
Log level
--config Configuration .ini file
--combinations Compute intersections for combinations of terms in query
--max-term-count,--mtc UINT Needs: --combinations
Max number of terms when computing combinations
--header Write TSV header
Description
Computes an intersection of posting lists given by the input queries.
It takes a file with queries and outputs the documents in the
intersection of the posting lists. See queries for
more details on the input parameters.
count-postings
Usage
Extracts posting counts from an inverted index.
Usage: ../../../build/bin/count-postings [OPTIONS]
Options:
-h,--help Print this help message and exit
-e,--encoding TEXT REQUIRED Index encoding
-i,--index TEXT REQUIRED Inverted index filename
--tokenizer TEXT:{english,whitespace} [english]
Tokenizer
-H,--html Strip HTML
-F,--token-filters TEXT:{krovetz,lowercase,porter2} ...
Token filters
--stopwords TEXT Path to file containing a list of stop words to filter out
-q,--queries TEXT Path to file with queries
--terms TEXT Term lexicon
--weighted Weights scores by query frequency
--sep TEXT Separator string
--query-id Print query ID at the beginning of each line, separated by a colon
-L,--log-level TEXT:{critical,debug,err,info,off,trace,warn} [info]
Log level
--config Configuration .ini file
--sum Sum postings accross the query terms; by default, individual list lengths will be printed, separated by the separator defined with --sep
Description
Extracts posting counts from an inverted index.
It sums up posting counts for each query term after parsing. See
parse_collection for more details about parsing options.
create_wand_data
Usage
Creates additional data for query processing.
Usage: ../../../build/bin/create_wand_data [OPTIONS]
Options:
-h,--help Print this help message and exit
-c,--collection TEXT REQUIRED
Collection basename
-o,--output TEXT REQUIRED Output filename
--quantize UINT Quantizes the scores using this many bits
--compress Needs: --quantize
Compress additional data
-s,--scorer TEXT REQUIRED Scorer function
--bm25-k1 FLOAT Needs: --scorer
BM25 k1 parameter.
--bm25-b FLOAT Needs: --scorer
BM25 b parameter.
--pl2-c FLOAT Needs: --scorer
PL2 c parameter.
--qld-mu FLOAT Needs: --scorer
QLD mu parameter.
--range Excludes: --block-size --lambda
Create docid-range based data
--terms-to-drop TEXT A filename containing a list of term IDs that we want to drop
-L,--log-level TEXT:{critical,debug,err,info,off,trace,warn} [info]
Log level
--config Configuration .ini file
[Option Group: blocks]
[At least 1 of the following options are required]
Options:
-b,--block-size UINT Excludes: --lambda --range
Block size for fixed-length blocks
-l,--lambda FLOAT Excludes: --block-size --range
Lambda parameter for variable blocks
Description
Creates additional data needed for certain query algorithms.
See "WAND" Data for more details.
Refer to queries for details about scoring functions.
Blocks
Each posting list is divided into blocks, and each block gets a
precomputed max score. These blocks can be either of equal size
throughout the index, defined by --block-size, or variable based on
the lambda parameter --lambda. [TODO: Explanation needed]
evaluate_queries
Usage
Retrieves query results in TREC format.
Usage: ../../../build/bin/evaluate_queries [OPTIONS]
Options:
-h,--help Print this help message and exit
-e,--encoding TEXT REQUIRED Index encoding
-i,--index TEXT REQUIRED Inverted index filename
-w,--wand TEXT REQUIRED WAND data filename
--compressed-wand Needs: --wand
Compressed WAND data file
--tokenizer TEXT:{english,whitespace} [english]
Tokenizer
-H,--html Strip HTML
-F,--token-filters TEXT:{krovetz,lowercase,porter2} ...
Token filters
--stopwords TEXT Path to file containing a list of stop words to filter out
-q,--queries TEXT Path to file with queries
--terms TEXT Term lexicon
--weighted Weights scores by query frequency
-k INT REQUIRED The number of top results to return
-a,--algorithm TEXT ... REQUIRED
Query processing algorithm
-s,--scorer TEXT REQUIRED Scorer function
--bm25-k1 FLOAT Needs: --scorer
BM25 k1 parameter.
--bm25-b FLOAT Needs: --scorer
BM25 b parameter.
--pl2-c FLOAT Needs: --scorer
PL2 c parameter.
--qld-mu FLOAT Needs: --scorer
QLD mu parameter.
-T,--thresholds TEXT File containing query thresholds
-j,--threads UINT Number of threads
-L,--log-level TEXT:{critical,debug,err,info,off,trace,warn} [info]
Log level
--config Configuration .ini file
-r,--run TEXT Run identifier
--documents TEXT REQUIRED Document lexicon
--quantized Quantized scores
Description
Returns results for the given queries. The results are printed in the
TREC format. See queries for detailed description of
the input parameters.
To print out the string identifiers of the documents (titles), you must
provide the document lexicon with --documents.
extract-maxscores
Usage
Extracts max-scores for query terms from an inverted index.
The max-scores will be printed to the output separated by --sep,
which is a tab by default.
Usage: ../../../build/bin/extract-maxscores [OPTIONS]
Options:
-h,--help Print this help message and exit
-w,--wand TEXT REQUIRED WAND data filename
--compressed-wand Needs: --wand
Compressed WAND data file
--tokenizer TEXT:{english,whitespace} [english]
Tokenizer
-H,--html Strip HTML
-F,--token-filters TEXT:{krovetz,lowercase,porter2} ...
Token filters
--stopwords TEXT Path to file containing a list of stop words to filter out
-q,--queries TEXT Path to file with queries
--terms TEXT Term lexicon
--weighted Weights scores by query frequency
--sep TEXT Separator string
--query-id Print query ID at the beginning of each line, separated by a colon
-L,--log-level TEXT:{critical,debug,err,info,off,trace,warn} [info]
Log level
--config Configuration .ini file
--quantized Quantized scores
extract_topics
Usage
A tool for converting queries from several formats to PISA queries.
Usage: ../../../build/bin/extract_topics [OPTIONS]
Options:
-h,--help Print this help message and exit
-L,--log-level TEXT:{critical,debug,err,info,off,trace,warn} [info]
Log level
--config Configuration .ini file
-i,--input TEXT REQUIRED TREC query input file
-o,--output TEXT REQUIRED Output basename
-f,--format TEXT REQUIRED Input format
-u,--unique Unique queries
invert
Usage
Constructs an inverted index from a forward index.
Usage: ../../../build/bin/invert [OPTIONS]
Options:
-h,--help Print this help message and exit
-i,--input TEXT REQUIRED Forward index basename
-o,--output TEXT REQUIRED Output inverted index basename
--term-count UINT Number of distinct terms in the forward index
-j,--threads UINT Number of threads
--batch-size UINT [100000] Number of documents to process at a time
-L,--log-level TEXT:{critical,debug,err,info,off,trace,warn} [info]
Log level
--config Configuration .ini file
kth_threshold
Usage
A tool for performing threshold estimation using the k-highest impact score for each term, pair or triple of a query. Pairs and triples are only used if provided with --pairs and --triples respectively.
Usage: ../../../build/bin/kth_threshold [OPTIONS]
Options:
-h,--help Print this help message and exit
-e,--encoding TEXT REQUIRED Index encoding
-i,--index TEXT REQUIRED Inverted index filename
-w,--wand TEXT REQUIRED WAND data filename
--compressed-wand Needs: --wand
Compressed WAND data file
--tokenizer TEXT:{english,whitespace} [english]
Tokenizer
-H,--html Strip HTML
-F,--token-filters TEXT:{krovetz,lowercase,porter2} ...
Token filters
--stopwords TEXT Path to file containing a list of stop words to filter out
-q,--queries TEXT Path to file with queries
--terms TEXT Term lexicon
--weighted Weights scores by query frequency
-k INT REQUIRED The number of top results to return
-s,--scorer TEXT REQUIRED Scorer function
--bm25-k1 FLOAT Needs: --scorer
BM25 k1 parameter.
--bm25-b FLOAT Needs: --scorer
BM25 b parameter.
--pl2-c FLOAT Needs: --scorer
PL2 c parameter.
--qld-mu FLOAT Needs: --scorer
QLD mu parameter.
-L,--log-level TEXT:{critical,debug,err,info,off,trace,warn} [info]
Log level
--config Configuration .ini file
-p,--pairs TEXT Excludes: --all-pairs
A tab separated file containing all the cached term pairs
-t,--triples TEXT Excludes: --all-triples
A tab separated file containing all the cached term triples
--all-pairs Excludes: --pairs
Consider all term pairs of a query
--all-triples Excludes: --triples
Consider all term triples of a query
--quantized Quantizes the scores
lexicon
Usage
Build, print, or query lexicon
Usage: ../../../build/bin/lexicon [OPTIONS] SUBCOMMAND
Options:
-h,--help Print this help message and exit
-L,--log-level TEXT:{critical,debug,err,info,off,trace,warn} [info]
Log level
--config Configuration .ini file
Subcommands:
build Build a lexicon
lookup Retrieve the payload at index
rlookup Retrieve the index of payload
print Print elements line by line
map_queries
Usage
A tool for transforming textual queries to IDs.
Usage: ../../../build/bin/map_queries [OPTIONS]
Options:
-h,--help Print this help message and exit
--tokenizer TEXT:{english,whitespace} [english]
Tokenizer
-H,--html Strip HTML
-F,--token-filters TEXT:{krovetz,lowercase,porter2} ...
Token filters
--stopwords TEXT Path to file containing a list of stop words to filter out
-q,--queries TEXT Path to file with queries
--terms TEXT Term lexicon
--weighted Weights scores by query frequency
--sep TEXT Separator string
--query-id Print query ID at the beginning of each line, separated by a colon
-L,--log-level TEXT:{critical,debug,err,info,off,trace,warn} [info]
Log level
--config Configuration .ini file
parse_collection
Usage
parse_collection - parse collection and store as forward index.
Usage: ../../../build/bin/parse_collection [OPTIONS] [SUBCOMMAND]
Options:
-h,--help Print this help message and exit
-L,--log-level TEXT:{critical,debug,err,info,off,trace,warn} [info]
Log level
-j,--threads UINT Number of threads
--tokenizer TEXT:{english,whitespace} [english]
Tokenizer
-H,--html Strip HTML
-F,--token-filters TEXT:{krovetz,lowercase,porter2} ...
Token filters
--stopwords TEXT Path to file containing a list of stop words to filter out
--config Configuration .ini file
-o,--output TEXT REQUIRED Forward index filename
-b,--batch-size INT [100000]
Number of documents to process in one thread
-f,--format TEXT [plaintext]
Input format
Subcommands:
merge Merge previously produced batch files. When parsing process was killed during merging, use this command to finish merging without having to restart building batches.
partition_fwd_index
Usage
Partition a forward index
Usage: ../../../build/bin/partition_fwd_index [OPTIONS]
Options:
-h,--help Print this help message and exit
-L,--log-level TEXT:{critical,debug,err,info,off,trace,warn} [info]
Log level
--config Configuration .ini file
-i,--input TEXT REQUIRED Forward index filename
-o,--output TEXT REQUIRED Basename of partitioned shards
-j,--threads INT Thread count
-r,--random-shards INT Excludes: --shard-files
Number of random shards
-s,--shard-files TEXT ... Excludes: --random-shards
List of files with shard titles
queries
Usage
Benchmarks queries on a given index.
Usage: ../../../build/bin/queries [OPTIONS]
Options:
-h,--help Print this help message and exit
-e,--encoding TEXT REQUIRED Index encoding
-i,--index TEXT REQUIRED Inverted index filename
-w,--wand TEXT WAND data filename
--compressed-wand Needs: --wand
Compressed WAND data file
--tokenizer TEXT:{english,whitespace} [english]
Tokenizer
-H,--html Strip HTML
-F,--token-filters TEXT:{krovetz,lowercase,porter2} ...
Token filters
--stopwords TEXT Path to file containing a list of stop words to filter out
-q,--queries TEXT Path to file with queries
--terms TEXT Term lexicon
--weighted Weights scores by query frequency
-k INT REQUIRED The number of top results to return
-a,--algorithm TEXT ... REQUIRED
Query processing algorithm
-s,--scorer TEXT REQUIRED Scorer function
--bm25-k1 FLOAT Needs: --scorer
BM25 k1 parameter.
--bm25-b FLOAT Needs: --scorer
BM25 b parameter.
--pl2-c FLOAT Needs: --scorer
PL2 c parameter.
--qld-mu FLOAT Needs: --scorer
QLD mu parameter.
-T,--thresholds TEXT File containing query thresholds
-L,--log-level TEXT:{critical,debug,err,info,off,trace,warn} [info]
Log level
--config Configuration .ini file
--quantized Quantized scores
--safe Needs: --thresholds Rerun if not enough results with pruning.
--runs UINT:POSITIVE [3] Number of runs per query
-o,--output TEXT Output file for per-run query timing data
Description
Runs query benchmarks focused on performance measurement, executing each query on the given index multiple times. Then, it aggregates statistics across all queries.
Note: for retrieval results use evaluate_queries.
Input
This program takes a compressed index as its input along with a file
containing the queries (line by line). Note that you need to specify the
correct index encoding with --encoding option, as this is currently
not stored in the index. If the index is quantized, you must pass
--quantized flag.
For certain types of retrieval algorithms, you will also need to pass the so-called "WAND file", which contains some metadata like skip lists and max scores.
Query Parsing
There are several parameters you can define to instruct the program on
how to parse and process the input queries, including which tokenizer to
use, whether to strip HTML from the query, and a list of token filters
(such as stemmers). For a more comprehensive description, see
parse_collection.
You can also pass a file containing stop-words, which will be excluded from the parsed queries.
In order for the parsing to actually take place, you need to also
provide the term lexicon with --terms. If not defined, the queries
will be interpreted as lists of document IDs.
Algorithm
You can specify what retrieval algorithm to use with --algorithm.
Furthermore, -k option defined how many results to retrieve for each
query.
Scoring
Use --scorer option to define which scoring function you want to use
(bm25, dph, pl2, qld). Some scoring functions have additional
parameters that you may override, see the help message above.
Thresholds
You can also pass a file with list of initial score thresholds. Any
documents that evaluate to a score below this value will be excluded.
This can speed up the algorithm, but if the threshold is too high, it
may exclude some of the relevant top-k results. If you want to always
ensure that the results are as if the initial threshold was zero, you
can pass --safe flag. It will force to recompute the entire query
without an initial threshold if it is detected that relevant documents
have been excluded. This may be useful if you have mostly accurate
threshold estimates, but still need the safety: even though some queries
will be slower, most will be much faster, thus improving overall
throughput and average latency.
read_collection
Usage
Reads binary collection to stdout.
Usage: ../../../build/bin/read_collection [OPTIONS] [SUBCOMMAND]
Options:
-h,--help Print this help message and exit
-L,--log-level TEXT:{critical,debug,err,info,off,trace,warn} [info]
Log level
--config Configuration .ini file
-c,--collection TEXT REQUIRED
Collection file path.
--maptext TEXT Excludes: --maplex
ID to string mapping in text file format. Line n is the string associated with ID n. E.g., if used to read a document from a forward index, this would be the `.terms` file, which maps term IDs to their string reperesentations.
--maplex TEXT Excludes: --maptext
ID to string mapping in lexicon binary file format. E.g., if used to read a document from a forward index, this would be the `.termlex` file, which maps term IDs to their string reperesentations.
Subcommands:
entry Reads single entry.
range Reads a range of entries.
reorder-docids
Usage
Reassigns the document IDs.
Usage: ../../../build/bin/reorder-docids [OPTIONS]
Options:
-h,--help Print this help message and exit
-c,--collection TEXT REQUIRED
Collection basename
-o,--output TEXT Output basename
--documents TEXT Document lexicon
--reordered-documents TEXT Needs: --documents
Reordered document lexicon
--seed UINT Needs: --random Random seed.
--store-fwdidx TEXT Needs: --recursive-graph-bisection
Output basename (forward index)
--fwdidx TEXT Needs: --recursive-graph-bisection
Use this forward index
-m,--min-len UINT Needs: --recursive-graph-bisection
Minimum list threshold
-d,--depth UINT:INT in [1 - 64] Needs: --recursive-graph-bisection Excludes: --node-config
Recursion depth
--node-config TEXT Needs: --recursive-graph-bisection Excludes: --depth
Node configuration file
--nogb Needs: --recursive-graph-bisection
No VarIntGB compression in forward index
-p,--print Needs: --recursive-graph-bisection
Print ordering to standard output
-j,--threads UINT Number of threads
-L,--log-level TEXT:{critical,debug,err,info,off,trace,warn} [info]
Log level
--config Configuration .ini file
[Option Group: methods]
[Exactly 1 of the following options is required]
Options:
--random Needs: --output Assign IDs randomly. You can use --seed for deterministic results.
--from-mapping TEXT Use the mapping defined in this new-line delimited text file
--by-feature TEXT Order by URLs from this file
--recursive-graph-bisection,--bp
Use recursive graph bisection algorithm
sample_inverted_index
Usage
A tool for sampling an inverted index.
Usage: ../../../build/bin/sample_inverted_index [OPTIONS]
Options:
-h,--help Print this help message and exit
-L,--log-level TEXT:{critical,debug,err,info,off,trace,warn} [info]
Log level
--config Configuration .ini file
-c,--collection TEXT REQUIRED
Input collection basename
-o,--output TEXT REQUIRED Output collection basename
-r,--rate FLOAT REQUIRED Sampling rate (proportional size of the output index)
-t,--type TEXT REQUIRED Sampling type
--terms-to-drop TEXT A filename containing a list of term IDs that we want to drop
--seed UINT Seed state
Description
Creates a smaller inverted index from an existing one by sampling postings or documents. The purpose of this tool is to reduce time and space requirements while preserving the main statistical properties of the original collection, making it useful for faster experiments and debugging.
Sampling strategy (-t, --type)
random_postings: keep random occurrences per posting list (not whole posting lists).random_docids: keep all postings belonging to a random subset of documents.
Examples
Keep ~25% of postings
sample_inverted_index \
-c path/to/inverted \
-o path/to/inverted.sampled \
-r 0.25 \
-t random_postings
Keep ~25% of the documents
sample_inverted_index \
-c path/to/inverted \
-o path/to/inverted.sampled \
-r 0.25 \
-t random_docids
selective_queries
Usage
Filters selective queries for a given index.
Usage: ../../../build/bin/selective_queries [OPTIONS]
Options:
-h,--help Print this help message and exit
-e,--encoding TEXT REQUIRED Index encoding
-i,--index TEXT REQUIRED Inverted index filename
--tokenizer TEXT:{english,whitespace} [english]
Tokenizer
-H,--html Strip HTML
-F,--token-filters TEXT:{krovetz,lowercase,porter2} ...
Token filters
--stopwords TEXT Path to file containing a list of stop words to filter out
-q,--queries TEXT Path to file with queries
--terms TEXT Term lexicon
--weighted Weights scores by query frequency
-L,--log-level TEXT:{critical,debug,err,info,off,trace,warn} [info]
Log level
--config Configuration .ini file
shards
Usage
Executes commands for shards.
Usage: ../../../build/bin/shards [OPTIONS] SUBCOMMAND
Options:
-h,--help Print this help message and exit
-L,--log-level TEXT:{critical,debug,err,info,off,trace,warn} [info]
Log level
--config Configuration .ini file
Subcommands:
invert Constructs an inverted index from a forward index.
reorder-docids Reorder document IDs.
compress Compresses an inverted index
wand-data Creates additional data for query processing.
taily-stats Extracts Taily statistics from the index and stores it in a file.
taily-score Computes Taily shard ranks for queries. NOTE: as term IDs need to be resolved individually for each shard, DO NOT provide already parsed and resolved queries (with IDs instead of terms).
taily-thresholds Computes Taily thresholds.
stem_queries
Usage
A tool for stemming PISA queries.
Usage: ../../../build/bin/stem_queries [OPTIONS]
Options:
-h,--help Print this help message and exit
-L,--log-level TEXT:{critical,debug,err,info,off,trace,warn} [info]
Log level
--config Configuration .ini file
-i,--input TEXT REQUIRED Query input file
-o,--output TEXT REQUIRED Query output file
--stemmer TEXT REQUIRED Stemmer
taily-stats
Usage
Extracts Taily statistics from the index and stores it in a file.
Usage: ../../../build/bin/taily-stats [OPTIONS]
Options:
-h,--help Print this help message and exit
-w,--wand TEXT REQUIRED WAND data filename
--compressed-wand Needs: --wand
Compressed WAND data file
-s,--scorer TEXT REQUIRED Scorer function
--bm25-k1 FLOAT Needs: --scorer
BM25 k1 parameter.
--bm25-b FLOAT Needs: --scorer
BM25 b parameter.
--pl2-c FLOAT Needs: --scorer
PL2 c parameter.
--qld-mu FLOAT Needs: --scorer
QLD mu parameter.
-L,--log-level TEXT:{critical,debug,err,info,off,trace,warn} [info]
Log level
-c,--collection TEXT REQUIRED
Binary collection basename
-o,--output TEXT REQUIRED Output file path
--config Configuration .ini file
taily-thresholds
Usage
Estimates query thresholds using Taily cut-offs.
Usage: ../../../build/bin/taily-thresholds [OPTIONS]
Options:
-h,--help Print this help message and exit
--tokenizer TEXT:{english,whitespace} [english]
Tokenizer
-H,--html Strip HTML
-F,--token-filters TEXT:{krovetz,lowercase,porter2} ...
Token filters
--stopwords TEXT Path to file containing a list of stop words to filter out
-q,--queries TEXT Path to file with queries
--terms TEXT Term lexicon
--weighted Weights scores by query frequency
-k INT REQUIRED The number of top results to return
-L,--log-level TEXT:{critical,debug,err,info,off,trace,warn} [info]
Log level
--stats TEXT REQUIRED Taily statistics file
--config Configuration .ini file
thresholds
Usage
Extracts query thresholds.
Usage: ../../../build/bin/thresholds [OPTIONS]
Options:
-h,--help Print this help message and exit
-e,--encoding TEXT REQUIRED Index encoding
-i,--index TEXT REQUIRED Inverted index filename
-w,--wand TEXT REQUIRED WAND data filename
--compressed-wand Needs: --wand
Compressed WAND data file
--tokenizer TEXT:{english,whitespace} [english]
Tokenizer
-H,--html Strip HTML
-F,--token-filters TEXT:{krovetz,lowercase,porter2} ...
Token filters
--stopwords TEXT Path to file containing a list of stop words to filter out
-q,--queries TEXT Path to file with queries
--terms TEXT Term lexicon
--weighted Weights scores by query frequency
-k INT REQUIRED The number of top results to return
-s,--scorer TEXT REQUIRED Scorer function
--bm25-k1 FLOAT Needs: --scorer
BM25 k1 parameter.
--bm25-b FLOAT Needs: --scorer
BM25 b parameter.
--pl2-c FLOAT Needs: --scorer
PL2 c parameter.
--qld-mu FLOAT Needs: --scorer
QLD mu parameter.
-L,--log-level TEXT:{critical,debug,err,info,off,trace,warn} [info]
Log level
--config Configuration .ini file
--quantized Quantizes the scores
Lookup Table Format Specification
A lookup table is a bidirectional mapping from an index, representing an
internal ID, to a binary payload, such as string. E.g., an N-element
lookup table maps values 0...N-1 to their payloads. These tables are
used for things like mapping terms to term IDs and document IDs to
titles or URLs.
The format of a lookup table is designed to operate without having to parse the entire structure. Once the header is parsed, it is possible to operate directly on the binary format to access the data. In fact, a lookup table will typically be memory mapped. Therefore, it is possible to perform a lookup (or reverse lookup) without loading the entire structure into memory.
The header always begins as follows:
+--------+--------+-------- -+
| 0x87 | Ver. | ... |
+--------+--------+-------- -+
The first byte is a constant identifier. When reading, we can verify whether this byte is correct to make sure we are using the correct type of data structure.
The second byte is equal to the version of the format.
The remaining of the format is defined separately for each version. The version is introduced in order to be able to update the format in the future but still be able to read old formats for backwards compatibility.
v1
+--------+--------+--------+--------+--------+--------+--------+--------+
| 0x87 | 0x01 | Flags | 0x00 |
+--------+--------+--------+--------+--------+--------+--------+--------+
| Length |
+--------+--------+--------+--------+--------+--------+--------+--------+
| |
| Offsets |
| |
+-----------------------------------------------------------------------+
| |
| Payloads |
| |
+-----------------------------------------------------------------------+
Immediately after the version bit, we have flags byte.
MSB LSB
+---+---+---+---+---+---+---+---+
| 0 | 0 | 0 | 0 | 0 | 0 | W | S |
+---+---+---+---+---+---+---+---+
The first bit (S) indicates whether the payloads are sorted (1) or not
(0). The second bit (W) defines the width of offsets (see below):
32-bit (0) or 64-bit (1). In most use cases, the cumulative size of the
payloads will be small enough to address it by 32-bit offsets. For
example, if we store words that are 16-bytes long on average, we can
address over 200 million of them. For this many elements, reducing the
width of the offsets would save us over 700 MB. Still, we want to
support 64-bit addressing because some payloads may be much longer
(e.g., URLs).
The rest of the bits in the flags byte are currently not used, but should be set to 0 to make sure that if more flags are introduced, we know what values to expect in the older iterations, and thus we can make sure to keep it backwards-compatible.
The following 5 bytes are padding with values of 0. This is to help with byte alignment. When loaded to memory, it should be loaded with 8-byte alignment. When memory mapped, it should be already correctly aligned by the operating system (at least on Linux).
Following the padding, there is a 64-bit unsigned integer encoding the
number of elements in the lexicon (N).
Given N and W, we can now calculate the byte range of all offsets,
and thus the address offset for the start of the payloads. The offsets
are N+1 little-endian unsigned integers of size determined by W
(either 4 or 8 bytes). The offsets are associated with consecutive IDs
from 0 to N-1; the last the N+1 offsets points at the first byte
after the last payload. The offsets are relative to the beginning of the
first payload, therefore the first offset will always be 0.
Payloads are arbitrary bytes, and must be interpreted by the software.
Although the typical use case are strings, this can be any binary
payload. Note that in case of strings, they will not be 0-terminated
unless they were specifically stored as such. Although this should be
clear by the fact a payload is simply a sequence of bytes, it is only
prudent to point it out. Thus, one must be extremely careful when using
C-style strings, as their use is contingent on a correct values inserted
and encoded in the first place, and assuming 0-terminated strings may
easily lead to undefined behavior. Thus, it is recommended to store
strings without terminating them, and then interpret them as string
views (such as std::string_view) instead of a C-style string.
The boundaries of the k-th payload are defined by the values of k-th and
(k+1)-th offsets. Note that because of the additional offset that points
to immediately after the last payload, we can read offsets k and k+1
for any index k < N (recall that N is the number of elements).
If the payloads are sorted (S), we can find an ID of a certain payload with a binary search. This is crucial for any application that requires mapping from payloads to their position in the table.